The state contract ¹ 02.435.11.1008 |
|
The validation of SOL docking program
|
|
Determination of positioning accuracy of ligands in proteins active sites This protocol concerns positioning accuracy of ligands in proteins active sites. The accuracy is defined by the root mean square deviation (RMSD) between ligand docked poses and experimental ligand poses taken from Protein Data Bank. The main difficulty of this validation is concerned with the selection of a suitable test set of protein-ligand complexes. We have selected protein-ligand complexes with respect to next requirements: high-quality experimental structures with high crystallographic resolution to exclude the presence of missing amino acids and atoms and diversity of ligands in selected protein-ligand complexes from small ones (several dozens of atoms) to large ones (more than one hundred atoms). We had no task to create original validation set, so the part of protein-ligand complexes were taken from the test sets described in.25,26 Metalloproteins were excluded from our set since the docking procedure for such complexes does not work correctly because of complexity of ligand-metal interaction calculation. Besides complexes containing different cofactors such as Heme, ATP, NADP and others were excluded as well. Finally we selected 80 complexes with the experimental structures taken directly from PDB27. The complexes were set up according to the rules described above (see the first protocol). Preliminary investigations of the docking procedure of program SOL have demonstrated that only 3 control parameters of 20 influence substantially on the docking success rate: the number of independent runs (NUMBER OF RUNS), population size or the number of individuals (POPULATION SIZE) and the number of generations participating in global optimization (NUMBER OF GENERATIONS). The values of these parameters during validation were set up: NUMBER OF RUNS: 50; POPULATION SIZE: 30000; NUMBER OF GENERATIONS: 500. For comparison AutoDock 3.05 program was used for the same test set with the same value of independent runs (NUMBER OF RUNS=50). The docking quality criterion for this type of validation protocol was the value of root mean square deviation (RMSD) between docked and experimental poses of native ligands taken from the respective PDB complexes. It is possible to distinguish four scales of docking quality: RMSD < 1 ? - excellent docking quality, 1 ? < RMSD < 2 ? - good quality, 2 ? < RMSD < 3 ? - satisfactory quality and 3 ? < RMSD – unsatisfactory quality. The results for 80 calculated complexes are presented in Table 2. Table 2. The PDB ID of calculated complexes, RMSD between docked and experimental positions of native ligands from these protein-ligand complexes obtained by SOL and AutoDock 3.05 programs, and the number of torsion degrees of freedom for heavy atoms of the respective ligands. The data were sorted according to increase of RMSD obtained by SOL.
Data presented in Table 2 show that the program SOL was able to dock with excellent and good quality (respective RMSD does not exceed 2 Å) 50 native ligands from 80 ones. Meanwhile the program AutoDock 3.05 has demonstrated such docking quality for 48 native ligands. Table 3 contains the number of complexes corresponding to each quality scale and their ratio to the total number of complexes in percents. Table 3.
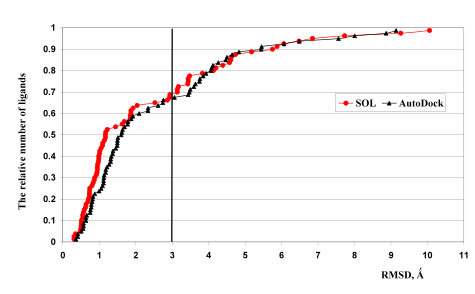
The results of the docking quality comparison for the both programs are illustrated by Fig.2. It demonstrates that the number of native ligands docked by SOL with RMSD ≤ 1 ? is almost two times larger than the respective number of ligands docked by AutoDock 3.05. The situation for 1 ? < RMSD < 2 ? is quite opposite. We did not observed correlation between docking quality demonstrated by SOL and AutoDock programs for the same complexes, i.e. docking quality (RMSD) for a given complex can be very different for SOL and AutoDock programs. For example, for complex 1ifu SOL gives RMSD equal to 0.63 Å, but RMSD obtained by AutoDock is equal to 8.87 Å. The docking quality of SOL is better than one of AutoDock, if we consider docking quality with the criterion RMSD < 1.5 Å. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||